New preprint on robust adaptation by single-celled organisms to environmental changes
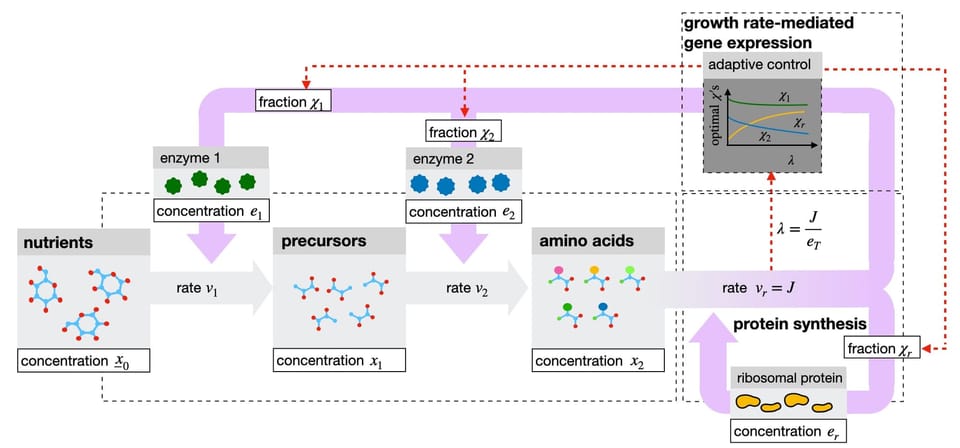
Single-celled organisms such as bacteria are able to tune enzyme levels that catalyze the reaction pathways by which they eventually make new copies of themselves. Depending on nutrient conditions, more or less enzyme is invested in different parts of their reaction network, so that reaction rates are constantly high, and cellular growth rate remains competitive. To adapt to changing conditions, microbes rely on gene regulatory networks that act on intracellular metabolic cues, which themselves are the result of the metabolic activity of the cell. Microbes thus have metabolic and gene networks that are coupled; as a combined whole they are able to adapt to changing conditions.
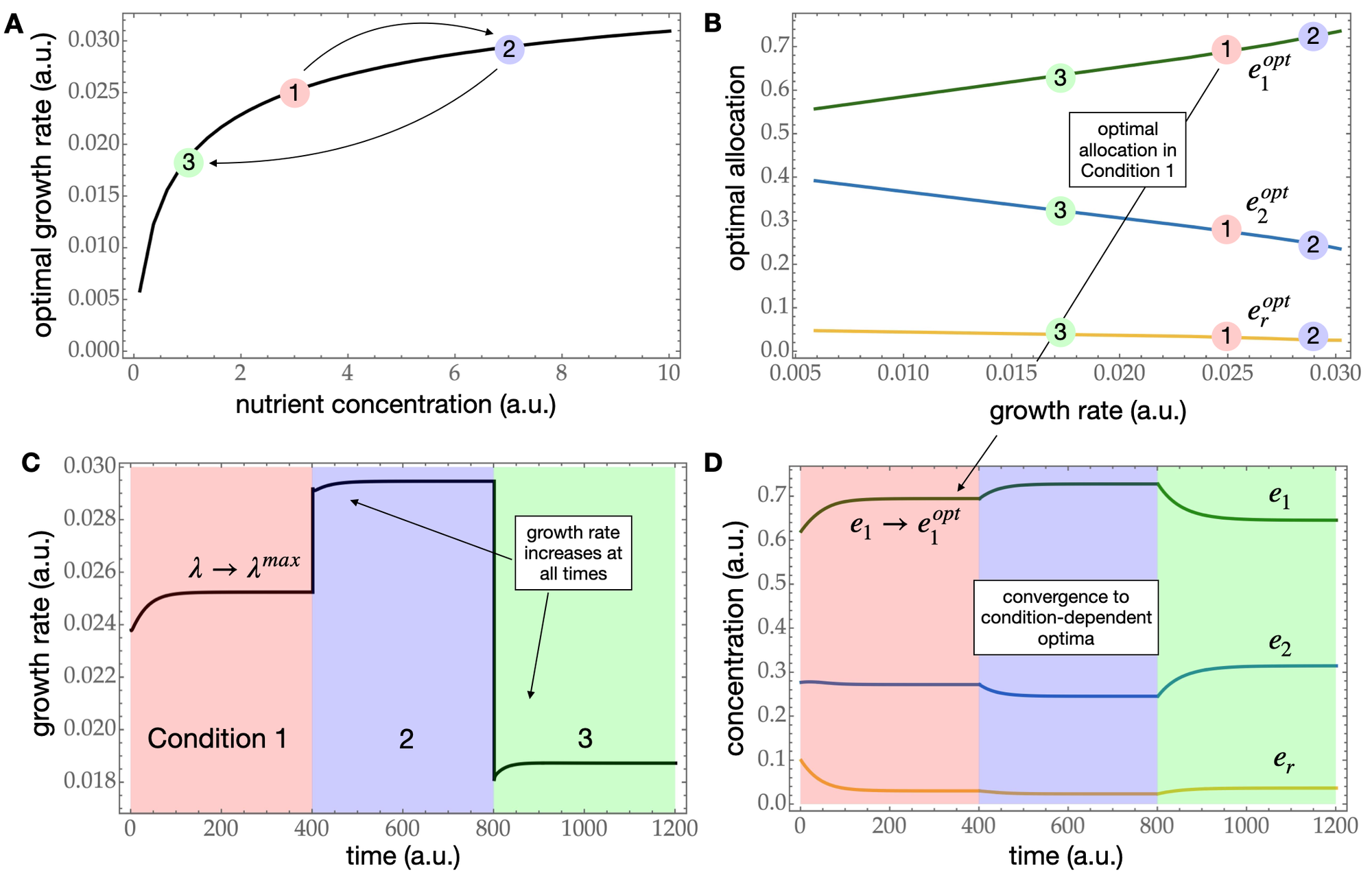
In this preprint, a collaboration between mathematicians Bob Planqué and Joost Hulshof and systems biologist Frank Bruggeman, it is shown that if microbes are able to sense their own growth rate (for instance, by a metabolite that faithfully covaries with it), they are able to always adapt and maximise their growth rate. The authors provide an explicit adaptive control with growth rate as input for the growth rate itself is shown to be a Lyapunov function. They then provide evidence that E. coli regulates its catabolic and anabolic enzyme pools in this way, indicating that microbes seem to able to implement this growth rate control strategy using simple biochemistry.